Contact us for publications not listed
Faculty Leader: Manish Patankar
Facility Manager: Dagna Sheerar
Recent Publications
Establishing a biosafety plan for a flow cytometry shared resource laboratory
A biosafety plan is essential to establish appropriate practices for biosafety in a shared resource laboratory (SRL). A biosafety plan will contain the essential information for the use of biological samples on specific instrumentation, their …
June 29, 2022Optimizing Flow Cytometric Analysis of Immune Cells in Samples Requiring Cryopreservation from Tumor-Bearing Mice
Most shared resource flow cytometry facilities do not permit analysis of radioactive samples. We are investigating low-dose molecular targeted radionuclide therapy (MTRT) as an immunomodulator in combination with in situ tumor vaccines and need to …
June 29, 2022- More Flow Publications
Scientific Accomplishments
Analytical Flow Cytometry Services
Response Monitoring in Clinical Research Trials
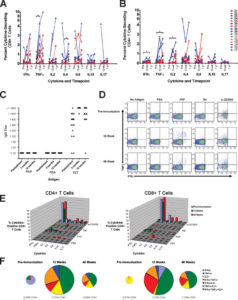
- FLOW supported the work of Douglas McNeel (DT) and colleagues in monitoring intracellular cytokine expression in T cells from patients receiving DNA vaccine encoding prostatic acid phosphatase using analytical flow cytometry
- FLOW provided real-time monitoring of immune response in patients recruited in a DNA vaccine clinical
- McNeel, DG. et al. Clin Can Res (2014) 20:3692-704
- R21 CA132267 and P30 CA014520 supplement

Cell Sorting Services
Supporting Translational Cancer Research
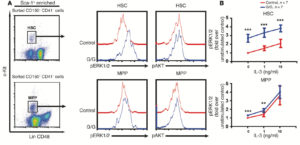
- FLOW supported the work of Jing Zhang (DT) and colleagues in monitoring MEK/ERK signaling in murine myeloproliferative neoplasms
- Hematopoietic Stem Cells and Myeloproliferative Progenitor Cells isolated by FACS and analyzed by flow cytometry for phosphorylation states of MEK and ERK pathways
- Kong, G., et al. J Clin Invest (2014) 124:276-273.
- 1R01CA152108 and 1R01HL113066

Imaging Cytometry Services
Supporting Basic Cancer Research
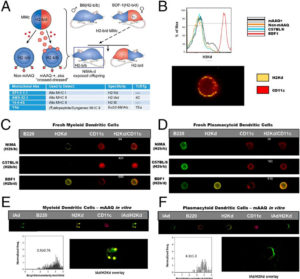
- FLOW supported the research of William Burlingham (TM) and colleagues in monitoring presentation of alloantigens on dendritic cells in murine microchimerism model to understand immune tolerance
- Dendritic Cells presenting antigens were detected and quantified by Imaging Cytometry and Flow Cytometry
- William Bracamonte-Baran et al. PNAS 2017;114:1099-1104
- 2R01-AI066219
