Selected scientific accomplishments from the SMSF are below. These include research from John Denu, Tim Bugni, David Andes, and Christian Capitini.
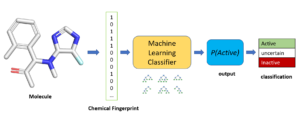
Machine Learning Models for Virtual Screening Ultra-Large Chemical Libraries
Anthony Gitter, PhD – Genetic and Epigenetic Mechanisms (GEM)
NIH NIGMS R01GM135631
- Given sufficient training data, simple machine learning models can be very effective in finding active molecules for new drug targets.
- Using prior screening data from SMSF, we trained a random forest classifier to predict compounds that inhibit the interaction between PriA helicase and the single-stranding binding protein. The model was tested prospectively on two large commercial chemical libraries.
- First, we virtually screened Aldrich Market Select chemical library (7 million molecules). Among the top compounds scored by the model, we purchased and tested 701 diverse candidates. Of these, 48% were active. In comparison, the hit rate in HTS was only 0.13%.
- Next, looking at performance on an ultra-large library, we applied the model to EnamineREAL (1.08 billion molecules). We purchased 68 diverse candidates prioritized by the model and observed activity in 31 compounds (46%), including one with an IC50 of 1.3 uM.
Alnammi M, Liu S, Ericksen SS, Ananiev GE, Voter AF, Guo S, Keck JL, Hoffmann FM, Wildman SA, Gitter A. ChemRxiv, 2021. doi:10.26434/chemrxiv-2021-fg8z9
Liu S, Alnammi M, Ericksen SS, Voter AF, Ananiev GE, Keck JL, Hoffmann FM, Wildman SA, Gitter A. J Chem Inf Model. 2019:59(1):282-293. doi: 10.1021/acs.jcim.8b00363.
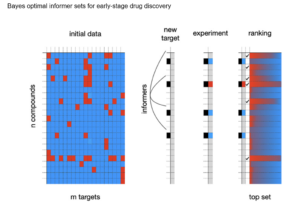
Optimal Probe Selection Strategies to Learn Ligand Selectivity of New Targets in a Target Class
Michael A. Newton, PhD – Genetic and Epigenetic Mechanisms (GEM)
NIH NIGMS R01GM135631
- What is the minimal compound testing data required to learn the broader ligand selectivity of some new drug target?
- Given a well-studied target class, we examined strategies for choosing small, information-rich probe sets or “informer sets” that, upon testing, provide key data to enable inference of a ligand selectivity profile for some new member of the target class.
- As a proof-of-concept, we developed and tested selection strategies that enable effective predictions of kinase inhibitor selectivity profiles on new kinase targets using probe sets of only 16 molecules.
- Excellent performance was observed both in retrospective tests on hundreds of human protein kinases and in prospective tests on 2 novel protein kinases from microbial pathogens.
Zhang H, Ericksen SS, Lee C-p, Ananiev GE, Wlodarchak N, Yu P, Mitchell JC, Gitter A, Wright SJ, Hoffmann FM, Wildman SA, Newton MA. PLOS Comp Biol. 2019. doi: 10.1371/journal.pcbi.1006813
Yu P, Ericksen SS, Gitter A, Newton MA. Biometrics 2022;1-3. doi: 10.1111/biom.13637
Discovery and Mechanism of Small Molecule Inhibitors Selective for the Chromatin-Binding Domains of Oncogenic UHRF1
John Denu, PhD – Genetic and Epigenetic Mechanisms (GEM)

NIH R01 GM059785
- Overexpressed UHRF1 has been observed in numerous cancers
- HTS of 100K compounds for inhibitors of UHRF1
- AlphaScreen in 1536 well plates
- Three lead compounds identified
Wallace H Liu, Robert E Miner 3rd, Brittany N Albaugh, Gene E Ananiev, Scott A Wildman, John M Denu. Biochemistry 2022 Mar 1;61(5):354-366. doi: 10.1021/acs.biochem.1c00698. Epub 2022 Feb 10
Biemamides A-E, Inhibitors of the TGF-β Pathway That Block the Epithelial to Mesenchymal Transition
Tim Bugni, PhD – Developmental Therapeutics Program (DT)

NIH R01 GM104192
- In established cancers, TGF-β is a central player in tumor growth, invasion, and subsequent metastasis.
- HTS of marine natural products for inhibitors of TGF-β.
- Five pyrimidinedione derivatives, biemamides A–E identified as inhibitors
- Biemamides blocked TGF-β-induced transition of the NMuMG epithelial cells into mesenchymal cells.
- Biemamides influence in vivo developmental processes related to body size regulation of Caenorhabditis elegans.
Zhang F, Braun DR, Ananiev GE, Hoffmann FM, Tsai IW, Rajski SR, Bugni TS. Org Lett. 2018 Sep 21;20(18):5529-5532. doi: 10.1021/acs.orglett.8b01871. Epub 2018 Aug 30.
A marine microbiome antifungal targets urgent-threat drug-resistant fungi
David Andes, MD and Tim Bugni, PhD – Infectious Diseases

U19 AI109673 and U19 AI142720
- A novel anti fungal agent Turbinmicin discovery from marine microbiome using MDR drug discovery platform.
- Turbinmicin displays potent in vitro and in vivo efficacy against multiple MDR fungal pathogens.
- MOA hypothesis is that turbinmicin impairs vesicle-mediated trafficking by inhibiting Sec14p.
Zhang F, Zhao M, Braun DR, Ericksen SS, Piotrowski JS, Nelson J, Peng J, Ananiev GE, Chanana S, Barns K, Fossen J, Sanchez H, Chevrette MG, Guzei IA, Zhao C, Guo L, Tang W, Currie CR, Rajski SR, Audhya A, Andes DR, Bugni TS. Science. 2020 Nov 20;370(6519):974-978. doi: 10.1126/science.abd6919.
Combining Immunocytokine and Ex Vivo Activated NK Cells as a Platform for Enhancing Graft-Versus-Tumor Effects Against GD2 + Murine Neuroblastoma
Christian Capitini, MD – Developmental Therapeutics Program (DT)
NCI/NIH R01 CA215461

- Management for high-risk neuroblastoma (NBL) has included autologous hematopoietic stem cell transplant (HSCT) and anti-GD2 immunotherapy, but survival remains around 50%.
- Allogeneic HSCT alone was insufficient to control NXS2 tumor growth, but the addition of hu14.18-IL2 controlled tumor growth and improved survival.
Bates PD, Rakhmilevich AL, Cho MM, Bouchlaka MN, Rao SL, Hales JM, Orentas RJ, Fry TJ, Gilles SD, Sondel PM, Capitini CM. Front Immunol. 2021 Aug 19;12:668307. doi: 10.3389/fimmu.2021.668307. eCollection 2021.
ACKNOWLEDGMENTS
Funding received from the UWCCC Cancer Center Support Grant (CCSG) for published research, including the use of UWCCC Shared Resources to generate or analyze data, or conduct clinical trials, needs to reference the UWCCC. See Acknowledging UWCCC in Publications, Posters, and Presentations. Please acknowledge “UWCCC Small Molecule Screening Facility” for any work supported by the SMSF. Thank you!
Sample Acknowledgment “The author(s) thank the University of Wisconsin Carbone Cancer Center Small Molecule Screening Facility, supported by P30 CA014520, for use of its facilities and services.”
View the UW Research Expertise website for more information on other UW–Madison laboratory facilities that provide state-of-the-art instrumentation and expert scientific staff.